Use Case: Highlighting Blood Cells in Dark-field Microscopy Using Image Segmentation
An application of dark-field microscopy is to view blood sample features such as blood cells. We created a use case for applying image segmentation to dark-field microscopy images using PerceptiLabs.
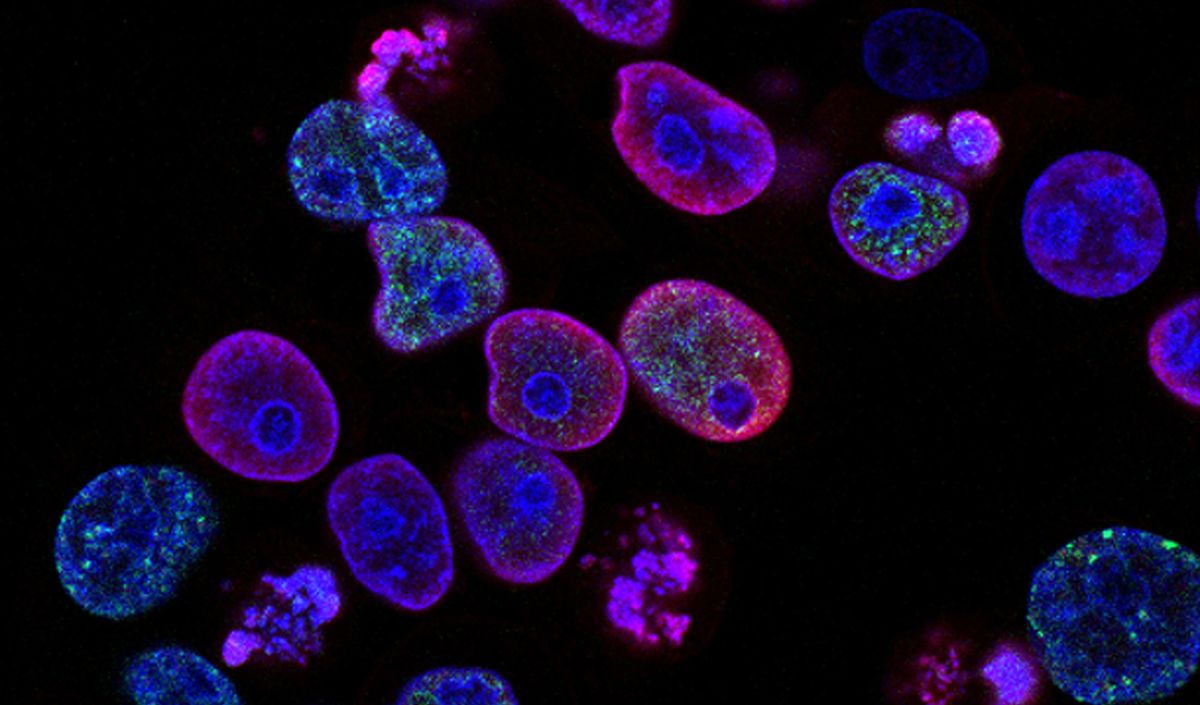
Dark-field microscopy is a microscopy technique that uses oblique illumination to enhance the contrast in specimens which are difficult to view under normal illumination. An interesting application of dark-field microscopy is to view blood sample features such as blood cells. Medical practitioners can identify and characterize patient samples, which can play an important role in diagnosing blood-related diseases.
Inspired by this, we created a use case for applying image segmentation to dark-field microscopy images using PerceptiLabs. Image segmentation is a powerful tool since it offers the capability to classify and highlight image features on a per-pixel level providing high granularity/better results. When applied to microscopy images of blood cells, such a model could potentially be used to alert medical and healthcare practitioners to the presence of viruses or bacteria.
Dataset
We started by looking for a set of dark-field microscopy images for blood cells and found them in this project1 on Kaggle that comprises 366 images.
The dataset also includes a set of masks which contain the ground truth segmentations on which to train the model. Figure 1 shows a few examples of these images:
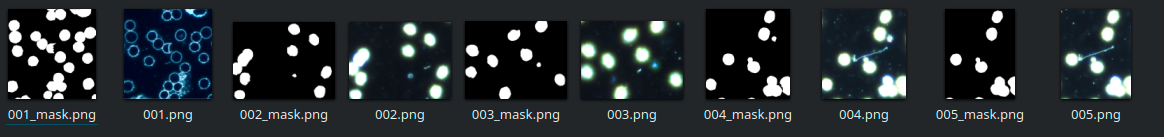
As of this writing, the U-Net component in PerceptiLabs only supports a single binary classification (i.e., it will only classify a single type of object/feature) but will support multiple classifications very soon. For single binary classification, PerceptiLabs requires masks to consist of values of 0 (black) for the background, and 255 (white) for the feature(s) of interest in the image. In order to use the masks from the dataset, we created a Jupyter Notebook which converts them to the required format.
The model was then trained and validated using the blood cell and converted mask images. To map the masks to their corresponding blood cell images, we created a .csv to load the data via PerceptiLabs' Data Wizard. Below is a partial example of how the .csv file looks:
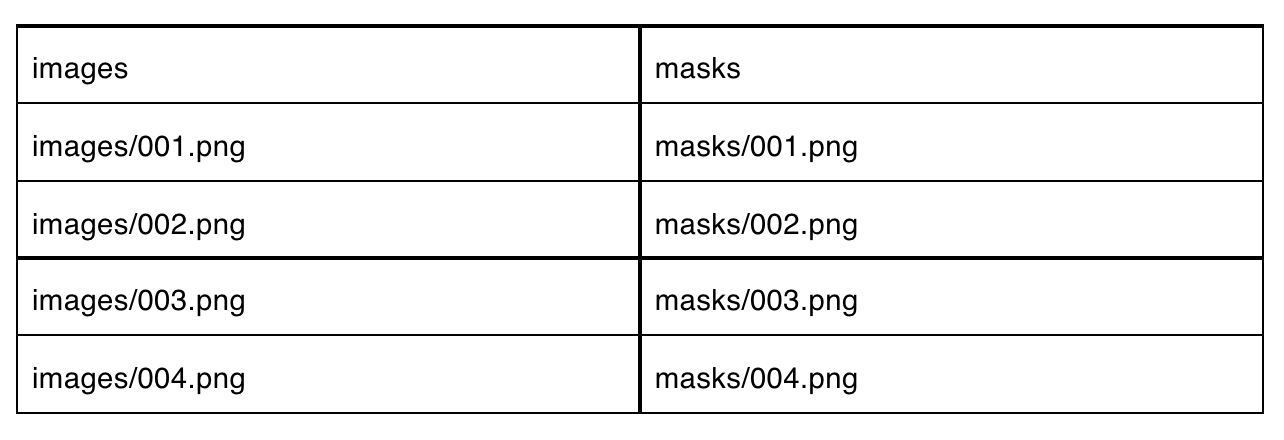
We've made the data and this CSV file available for experimentation on GitHub.
Model Summary
Our model was built with just one Component, a U-Net configured with VGG16 as its backbone, pretrained with ImageNet:
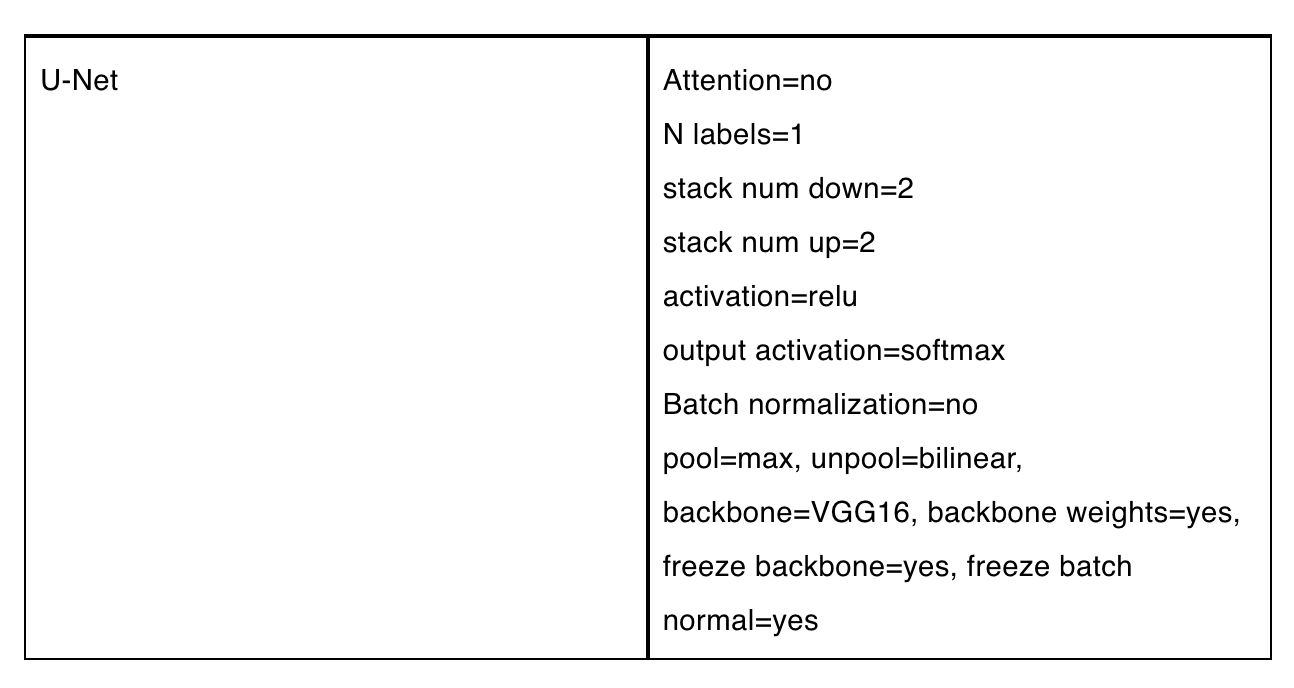
Figure 2 shows the model's topology in PerceptiLabs:
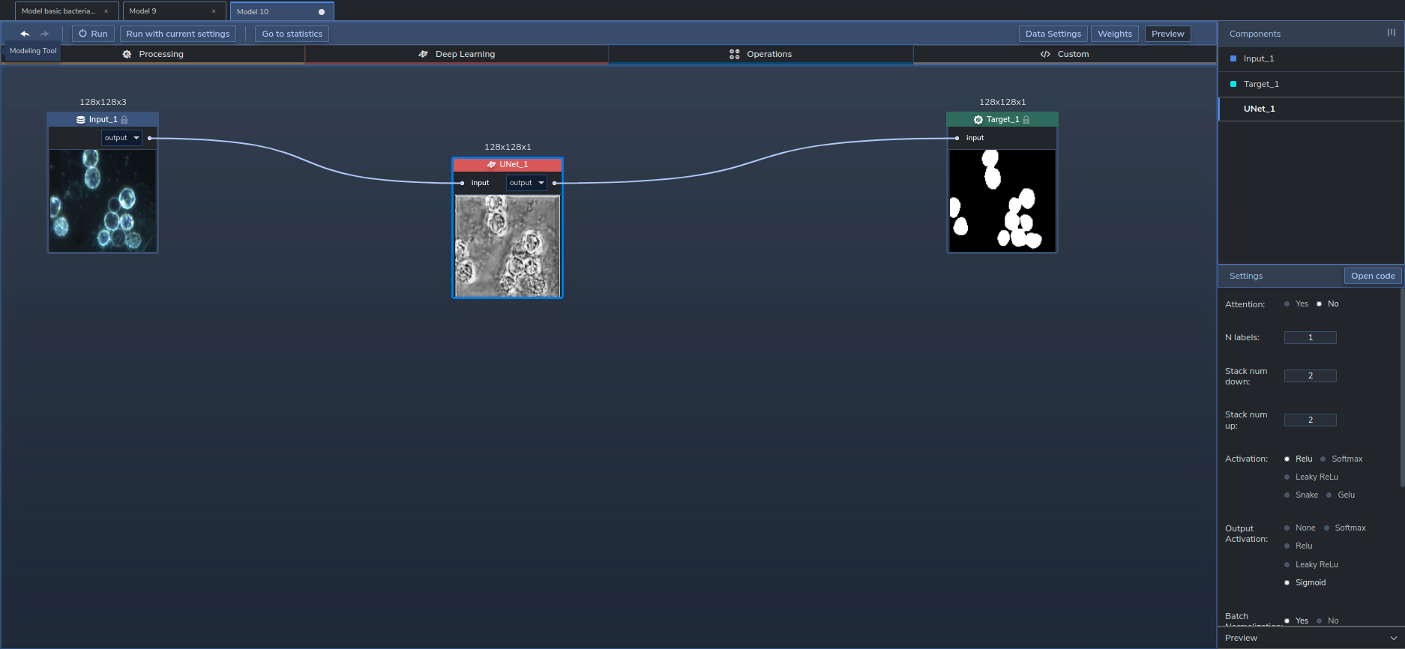
We chose to use a U-Net because of its strengths in segmenting images, while its VGG16 backbone is known to provide high-performing convolutions. And since the VGG has been pre-trained, using those existing weights results in faster training times.
Training and Results
We trained the model with 10 epochs in batches of 32, using the ADAM optimizer, a learning rate of 0.001, and a Dice loss function.
Since the output of this model comprises segmentations (i.e., segmented regions of images representing blood cells), the best method to assess the model's accuracy is to compare the output against the ground truth segmentation in the mask images using Intersection over Union (IoU). With a training time of around 131 seconds, we were able to achieve a training IoU of 0.86 and a validation IoU of 0.81%. Note that IoU values over 0.5 are generally considered good by the ML community2. Figure 3 shows the results after training:
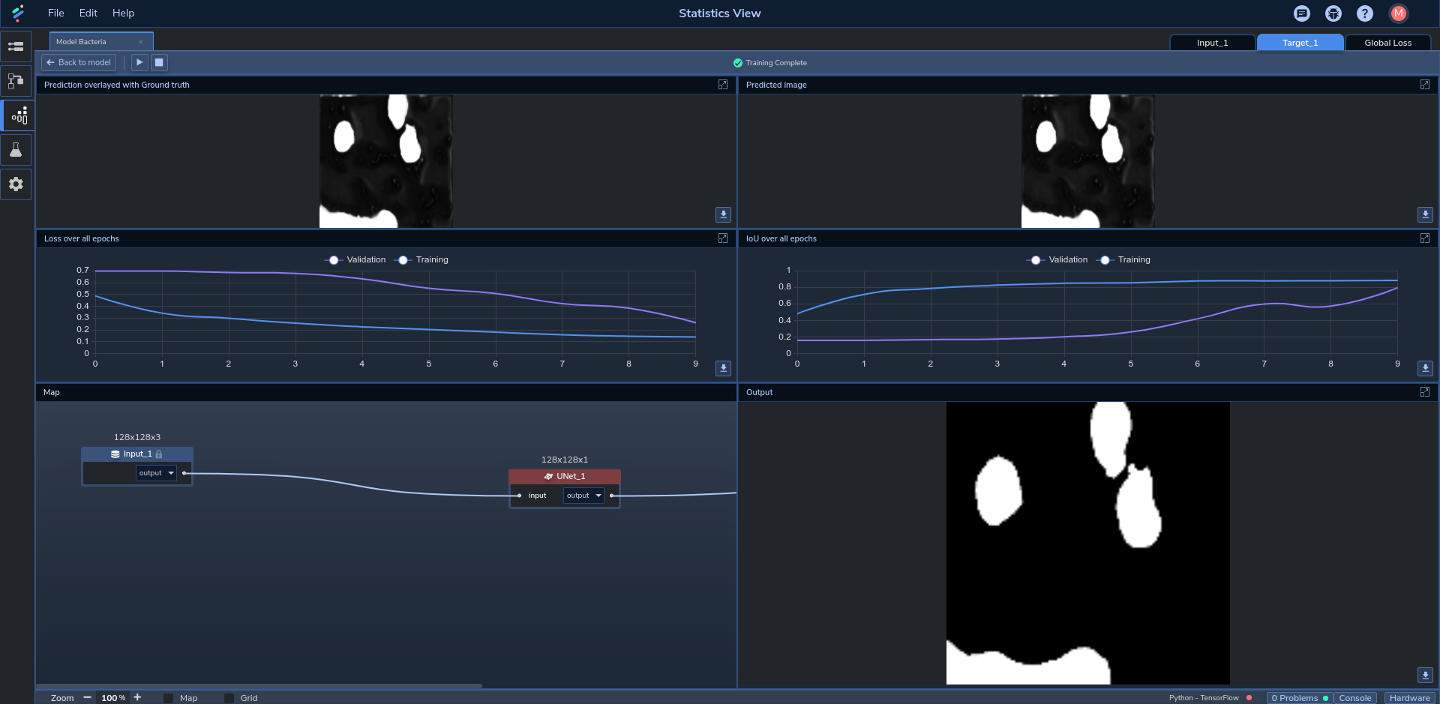
In the top-left pane we can see an image overlaid with its corresponding (ground truth) mask. In the middle-right pane we can see that the training IoU started at just under 0.5 and quickly ramped up to past 0.8 over the first two epochs before gradually stabilizing. Validation IoU, on the other hand, remained fairly low until about the fifth epoch, after which it ramped up to around 0.8 by the end of the last epoch. In the middle left pane we can see that training loss started at around 0.5 and gradually decreased to just over 0.1 by the final epoch. Validation loss remained high at over 0.7 through to around the fourth epoch before decreasing to just over 0.2 by the end of the final epoch.
Vertical Applications
A model like this could be used as the basis for creating new models that can segment other types of images. Such models could be applied across a range of verticals including other medical/healthcare use cases (e.g., x-rays), automotive (e.g., computer vision for autonomous vehicles), security (e.g., classifying people in camera footage), and IoT (e.g., classifying objects on an assembly line).
Summary
Image segmentation is a powerful tool for classifying features on a per-pixel level. The model in this use case is an example of how image segmentation can be applied for medical/healthcare applications, to help practitioners more quickly and easily identify different types of blood cells in dark-field microscopy images.
If you want to build a deep learning model similar to this, run PerceptiLabs and grab a copy of our pre-processed dataset from GitHub.
1Long Pollehn, Bacteria detection with darkfield microscopy – Dataset for spirochaeta segmentation with image and manually annotated masks, 2020, Kaggle.com, https://www.kaggle.com/longnguyen2306/bacteria-detection-with-darkfield-microscopy, Data files © Original Authors
2https://www.pyimagesearch.com/2016/11/07/intersection-over-union-iou-for-object-detection/